Ecology and Environment ›› 2023, Vol. 32 ›› Issue (5): 989-1000.DOI: 10.16258/j.cnki.1674-5906.2023.05.016
• Research Articles • Previous Articles Next Articles
KOU Zhu1,2( ), QING Chun1,2, YUAN Changguo1,2, LI Ping1,2,*(
), QING Chun1,2, YUAN Changguo1,2, LI Ping1,2,*( )
)
Received:2023-02-27
Online:2023-05-18
Published:2023-08-09
Contact:
LI Ping
寇祝1,2( ), 卿纯1,2, 袁昌果1,2, 李平1,2,*(
), 卿纯1,2, 袁昌果1,2, 李平1,2,*( )
)
通讯作者:
李平
作者简介:寇祝(1997年生),女,硕士研究生,主要从事环境微生物方面的研究。E-mail: kouzhu_kz@163.com
基金资助:CLC Number:
KOU Zhu, QING Chun, YUAN Changguo, LI Ping. Diversity and Distribution of Sulfur Oxidizing Bacteria in Hot Springs of Northeast Tibet, China[J]. Ecology and Environment, 2023, 32(5): 989-1000.
寇祝, 卿纯, 袁昌果, 李平. 西藏东北部热泉水中硫氧化菌的多样性及分布特征[J]. 生态环境学报, 2023, 32(5): 989-1000.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.jeesci.com/EN/10.16258/j.cnki.1674-5906.2023.05.016
| 采样点 | 温度 t/℃ | pH | 氧化还原电位 ORP/mV | ρ(五价砷As5+)/ (µg·L-1) | ρ(总砷AsT)/ (µg·L-1) | ρ(硫酸根SO42-)/ (mg·L-1) | ρ(硫离子S2-)/ (mg·L-1) | ρ(总铁FeT)/ (mg·L-1) | ρ(碳酸氢根HCO3-)/ (mg·L-1) |
|---|---|---|---|---|---|---|---|---|---|
| DB1 | 56.0±0.1e1) | 7.19±0.10g | -253.4±0.3g | 8331.27±2.02b | 8331.27±2.02b | 615.1±1.5c | 1.92±0.03e | 0.010±0.006h | 2221.08±1.35a |
| DB41 | 69.8±0.2a | 7.27±0.15e | -301.0±0.4j | 9298.58±1.08a | 9298.58±1.08a | 643.3±0.9a | 1.55±0.04j | 0.057±0.025g | ND |
| DB45 | 37.7±0.1i | 8.37±0.01a | -105.8±0.5b | 2891.80±1.15f | 2891.80±1.15g | 455.6±2.1h | 1.52±0.01k | ND2) | 1763.48±1.95c |
| DB5 | 51.4±0.1f | 7.21±0.15fg | -157.5±0.8e | 1279.50±1.37h | 6053.57±1.10d | 557.3±1.4f | 2.00±0.03d | 0.186±0.002e | 1434.05±1.11d |
| DB6 | 50.2±0.1g | 7.01±0.01j | -307.3±0.5k | 7415.97±1.54c | 7415.97±1.54c | 604.2±0.9d | 1.76±0.02g | 0.144±0.002f | 1385.16±1.83e |
| DB7 | 36.7±0.2j | 7.35±0.02d | -126.6±0.3d | 2331.15±1.06g | 2331.15±1.06h | 536.1±1.8g | 1.62±0.02i | 0.495±0.003c | 2105.17±1.33b |
| DB8 | 28.5±0.2l | 8.01±0.01b | -109.9±0.3c | 4211.16±2.22d | 4211.16±2.22e | 578.1±1.4e | 1.67±0.03h | 0.200±0.002d | 1113.62±1.99h |
| DB10 | 35.0±0.1k | 7.05±0.04i | -229.5±0.4f | 3445.68±0.97e | 3445.68±0.97f | 47.7±1.0l | 2.08±0.02c | 0.144±0.003f | 1354.56±1.93f |
| ZM1 | 69.2±0.1b | 7.11±0.03h | -301.3±0.4j | 24.57±2.32k | 29.27±2.33l | 54.4±1.1k | 2.56±0.02a | 0.495±0.003c | 1113.60±1.71h |
| ZM2 | 61.0±0.0d | 7.66±0.03c | -277.3±0.8h | ND | ND | 31.7±1.2m | 2.16±0.03b | 0.200±0.001d | 988.50±2.28j |
| BDW | 38.2±0.1h | 6.59±0.01k | -45.5±0.3a | 0.00±0.00l | 43.17±2.08k | 620.7±1.3b | 0.80±0.04l | 3.315±0.003b | 1025.11±1.55i |
| QS | 66.0±0.0c | 7.23±0.01f | -282.5±0.4i | 235.89±2.00j | 235.89±2.00j | 125.4±1.5i | 1.82±0.02f | 0.183±0.002e | 1189.93±0.87g |
| GN | 25.0±0.1m | 7.07±0.01i | ND | 346.66±1.51i | 346.66±1.51i | 109.6±1.8j | ND | 4.294±0.005a | ND |
Table 1 Main geochemical parameters of hot spring water in Tibet
| 采样点 | 温度 t/℃ | pH | 氧化还原电位 ORP/mV | ρ(五价砷As5+)/ (µg·L-1) | ρ(总砷AsT)/ (µg·L-1) | ρ(硫酸根SO42-)/ (mg·L-1) | ρ(硫离子S2-)/ (mg·L-1) | ρ(总铁FeT)/ (mg·L-1) | ρ(碳酸氢根HCO3-)/ (mg·L-1) |
|---|---|---|---|---|---|---|---|---|---|
| DB1 | 56.0±0.1e1) | 7.19±0.10g | -253.4±0.3g | 8331.27±2.02b | 8331.27±2.02b | 615.1±1.5c | 1.92±0.03e | 0.010±0.006h | 2221.08±1.35a |
| DB41 | 69.8±0.2a | 7.27±0.15e | -301.0±0.4j | 9298.58±1.08a | 9298.58±1.08a | 643.3±0.9a | 1.55±0.04j | 0.057±0.025g | ND |
| DB45 | 37.7±0.1i | 8.37±0.01a | -105.8±0.5b | 2891.80±1.15f | 2891.80±1.15g | 455.6±2.1h | 1.52±0.01k | ND2) | 1763.48±1.95c |
| DB5 | 51.4±0.1f | 7.21±0.15fg | -157.5±0.8e | 1279.50±1.37h | 6053.57±1.10d | 557.3±1.4f | 2.00±0.03d | 0.186±0.002e | 1434.05±1.11d |
| DB6 | 50.2±0.1g | 7.01±0.01j | -307.3±0.5k | 7415.97±1.54c | 7415.97±1.54c | 604.2±0.9d | 1.76±0.02g | 0.144±0.002f | 1385.16±1.83e |
| DB7 | 36.7±0.2j | 7.35±0.02d | -126.6±0.3d | 2331.15±1.06g | 2331.15±1.06h | 536.1±1.8g | 1.62±0.02i | 0.495±0.003c | 2105.17±1.33b |
| DB8 | 28.5±0.2l | 8.01±0.01b | -109.9±0.3c | 4211.16±2.22d | 4211.16±2.22e | 578.1±1.4e | 1.67±0.03h | 0.200±0.002d | 1113.62±1.99h |
| DB10 | 35.0±0.1k | 7.05±0.04i | -229.5±0.4f | 3445.68±0.97e | 3445.68±0.97f | 47.7±1.0l | 2.08±0.02c | 0.144±0.003f | 1354.56±1.93f |
| ZM1 | 69.2±0.1b | 7.11±0.03h | -301.3±0.4j | 24.57±2.32k | 29.27±2.33l | 54.4±1.1k | 2.56±0.02a | 0.495±0.003c | 1113.60±1.71h |
| ZM2 | 61.0±0.0d | 7.66±0.03c | -277.3±0.8h | ND | ND | 31.7±1.2m | 2.16±0.03b | 0.200±0.001d | 988.50±2.28j |
| BDW | 38.2±0.1h | 6.59±0.01k | -45.5±0.3a | 0.00±0.00l | 43.17±2.08k | 620.7±1.3b | 0.80±0.04l | 3.315±0.003b | 1025.11±1.55i |
| QS | 66.0±0.0c | 7.23±0.01f | -282.5±0.4i | 235.89±2.00j | 235.89±2.00j | 125.4±1.5i | 1.82±0.02f | 0.183±0.002e | 1189.93±0.87g |
| GN | 25.0±0.1m | 7.07±0.01i | ND | 346.66±1.51i | 346.66±1.51i | 109.6±1.8j | ND | 4.294±0.005a | ND |
| 采样点 | 序列数 | OTU数 | 覆盖度/ % | Chao 1 指数 | Shannon指数 | Simpson_ 1-D指数 |
|---|---|---|---|---|---|---|
| DB1 | 25 | 4 | 96 | 4 | 1.4 | 0.5 |
| DB45 | 16 | 5 | 94 | 5 | 2.1 | 0.7 |
| DB5 | 23 | 5 | 96 | 5 | 2.0 | 0.7 |
| DB6 | 25 | 3 | 96 | 3 | 1.0 | 0.4 |
| DB7 | 22 | 6 | 95 | 6 | 2.4 | 0.8 |
| DB8 | 17 | 4 | 100 | 4 | 1.9 | 0.7 |
| DB10 | 29 | 4 | 93 | 5 | 1.2 | 0.5 |
| BDW | 23 | 4 | 95 | 4 | 1.2 | 0.4 |
| QS | 27 | 5 | 92 | 6 | 1.6 | 0.6 |
| GN | 20 | 7 | 95 | 7 | 2.6 | 0.8 |
Table 2 Diversity indices of dsrA gene clone libraries
| 采样点 | 序列数 | OTU数 | 覆盖度/ % | Chao 1 指数 | Shannon指数 | Simpson_ 1-D指数 |
|---|---|---|---|---|---|---|
| DB1 | 25 | 4 | 96 | 4 | 1.4 | 0.5 |
| DB45 | 16 | 5 | 94 | 5 | 2.1 | 0.7 |
| DB5 | 23 | 5 | 96 | 5 | 2.0 | 0.7 |
| DB6 | 25 | 3 | 96 | 3 | 1.0 | 0.4 |
| DB7 | 22 | 6 | 95 | 6 | 2.4 | 0.8 |
| DB8 | 17 | 4 | 100 | 4 | 1.9 | 0.7 |
| DB10 | 29 | 4 | 93 | 5 | 1.2 | 0.5 |
| BDW | 23 | 4 | 95 | 4 | 1.2 | 0.4 |
| QS | 27 | 5 | 92 | 6 | 1.6 | 0.6 |
| GN | 20 | 7 | 95 | 7 | 2.6 | 0.8 |
| 采样点 | 序列数 | OTU数 | 覆盖度/ % | Chao 1指数 | Shannon指数 | Simpson_ 1-D指数 |
|---|---|---|---|---|---|---|
| DB1 | 20 | 3 | 95 | 3.0 | 1.1 | 0.4 |
| DB41 | 41 | 8 | 95 | 12.0 | 2.9 | 0.8 |
| DB5 | 41 | 5 | 97 | 5.5 | 1.9 | 0.7 |
| DB6 | 40 | 8 | 95 | 9.0 | 2.3 | 0.7 |
| DB7 | 37 | 7 | 94 | 8.0 | 2.2 | 0.7 |
| DB8 | 28 | 9 | 93 | 10.0 | 2.9 | 0.8 |
| DB10 | 38 | 6 | 95 | 7.0 | 1.6 | 0.5 |
| ZM1 | 25 | 7 | 92 | 6.5 | 2.2 | 0.7 |
| ZM2 | 24 | 4 | 92 | 5.0 | 0.9 | 0.3 |
| BDW | 20 | 4 | 95 | 4.5 | 1.5 | 0.6 |
Table 3 Diversity indices of soxB gene clone libraries
| 采样点 | 序列数 | OTU数 | 覆盖度/ % | Chao 1指数 | Shannon指数 | Simpson_ 1-D指数 |
|---|---|---|---|---|---|---|
| DB1 | 20 | 3 | 95 | 3.0 | 1.1 | 0.4 |
| DB41 | 41 | 8 | 95 | 12.0 | 2.9 | 0.8 |
| DB5 | 41 | 5 | 97 | 5.5 | 1.9 | 0.7 |
| DB6 | 40 | 8 | 95 | 9.0 | 2.3 | 0.7 |
| DB7 | 37 | 7 | 94 | 8.0 | 2.2 | 0.7 |
| DB8 | 28 | 9 | 93 | 10.0 | 2.9 | 0.8 |
| DB10 | 38 | 6 | 95 | 7.0 | 1.6 | 0.5 |
| ZM1 | 25 | 7 | 92 | 6.5 | 2.2 | 0.7 |
| ZM2 | 24 | 4 | 92 | 5.0 | 0.9 | 0.3 |
| BDW | 20 | 4 | 95 | 4.5 | 1.5 | 0.6 |
| 环境 因子 | Chao1指数 | Shannon指数 | Simpson_1-D指数 | |||||
|---|---|---|---|---|---|---|---|---|
| r | P | r | P | r | P | |||
| pH | 0.244 | 0.497 | 0.591 | 0.072 | 0.642 | 0.045 | ||
| ORP | 0.280 | 0.432 | 0.289 | 0.418 | 0.327 | 0.356 | ||
| As5+ | -0.476 | 0.165 | -0.154 | 0.671 | -0.099 | 0.786 | ||
| T | -0.500 | 0.141 | -0.474 | 0.166 | -0.543 | 0.105 | ||
| AsT | -0.530 | 0.115 | -0.098 | 0.787 | -0.105 | 0.773 | ||
| SO42- | -0.854 | 0.002 | -0.455 | 0.186 | -0.451 | 0.191 | ||
| S2- | -0.177 | 0.625 | -0.302 | 0.397 | -0.358 | 0.310 | ||
| FeT | 0.468 | 0.173 | 0.457 | 0.184 | 0.433 | 0.211 | ||
| HCO3- | -0.146 | 0.687 | 0.068 | 0.853 | 0.012 | 0.973 | ||
Table 4 Correlation analysis between environmental factors and alpha diversity index of dsrA
| 环境 因子 | Chao1指数 | Shannon指数 | Simpson_1-D指数 | |||||
|---|---|---|---|---|---|---|---|---|
| r | P | r | P | r | P | |||
| pH | 0.244 | 0.497 | 0.591 | 0.072 | 0.642 | 0.045 | ||
| ORP | 0.280 | 0.432 | 0.289 | 0.418 | 0.327 | 0.356 | ||
| As5+ | -0.476 | 0.165 | -0.154 | 0.671 | -0.099 | 0.786 | ||
| T | -0.500 | 0.141 | -0.474 | 0.166 | -0.543 | 0.105 | ||
| AsT | -0.530 | 0.115 | -0.098 | 0.787 | -0.105 | 0.773 | ||
| SO42- | -0.854 | 0.002 | -0.455 | 0.186 | -0.451 | 0.191 | ||
| S2- | -0.177 | 0.625 | -0.302 | 0.397 | -0.358 | 0.310 | ||
| FeT | 0.468 | 0.173 | 0.457 | 0.184 | 0.433 | 0.211 | ||
| HCO3- | -0.146 | 0.687 | 0.068 | 0.853 | 0.012 | 0.973 | ||

Figure 3 Neighbor-joining tree showing the phylogenetic relationships of the deduced soxB amino acid sequences translated from the soxB gene OTU clone sequences
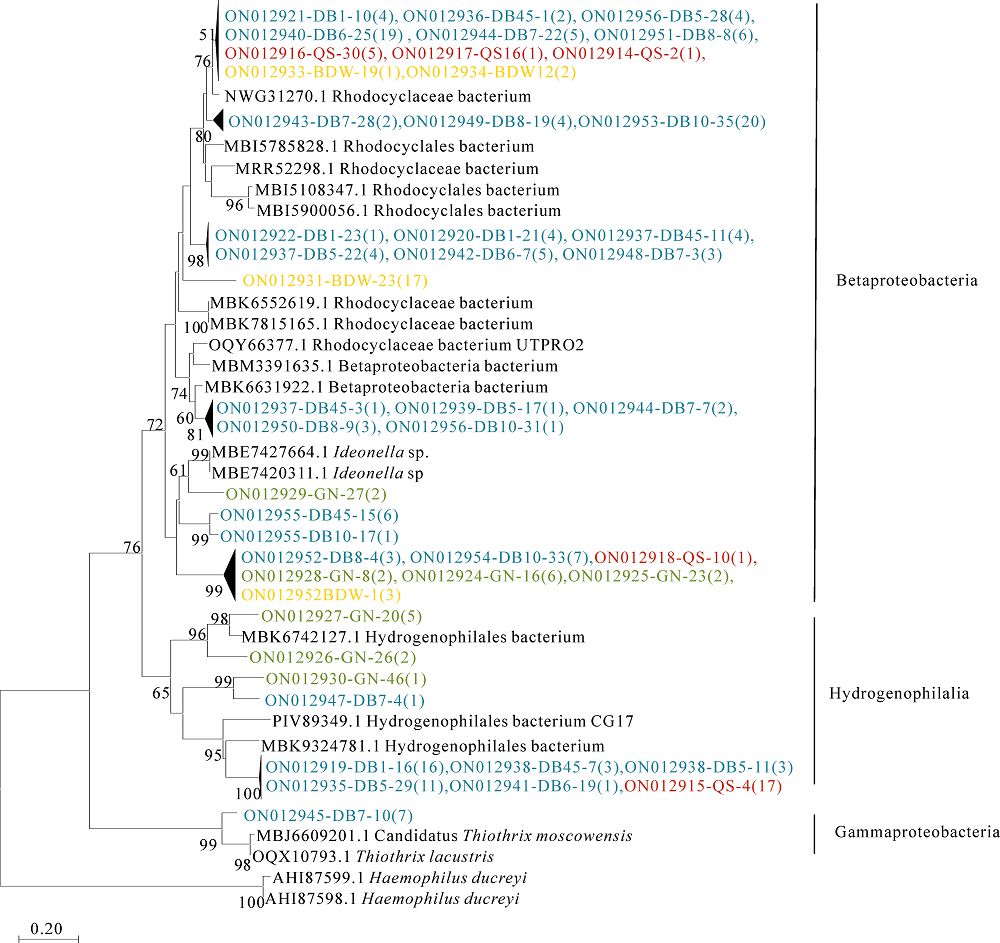
Figure 4 Neighbor-joining tree showing the phylogenetic relationships of the deduced dsrA amino acid sequences translated from the dsrA gene OTU clone sequences
| 样点 名称 | soxB基因拷贝数/ (copies·mL-1) | dsrA基因拷贝数/ (copies·mL-1) | 16S rRNA基因拷贝数/ (copies·mL-1) | soxB基因相对丰度/ % | dsrA基因相对丰度/ % | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| soxB | SD | dsrA | SD | 16S2) | SD | soxB/16S | dsrA/16S | ||||
| DB1 | 4.53E+04 | 1.16E+04 | 2.24E+05 | 2.83E+03 | 7.41E+05 | 4.22E+05 | 6.12 | 30.25 | |||
| DB41 | 3.62E+03 | 7.85E+02 | ND | ND | 2.31E+05 | 4.88E+04 | 1.57 | ND | |||
| DB45 | ND1) | ND | 2.18E+02 | 1.48E+02 | 9.69E+02 | 6.36E+00 | ND | 22.50 | |||
| DB5 | 2.14E+05 | 3.61E+03 | 8.42E+04 | 3.99E+03 | 4.66E+06 | 2.51E+05 | 4.59 | 1.80 | |||
| DB6 | 3.72E+03 | 1.12E+03 | 2.07E+04 | 1.59E+03 | 2.50E+05 | 7.35E+04 | 1.49 | 8.28 | |||
| DB7 | 1.37E+05 | 3.17E+04 | 6.55E+04 | 7.06E+03 | 3.43E+06 | 4.24E+04 | 4.00 | 1.91 | |||
| DB8 | 4.89E+04 | 1.07E+04 | 1.36E+04 | 1.46E+03 | 2.23E+06 | 1.06E+05 | 2.19 | 0.61 | |||
| DB10 | 3.31E+04 | 2.25E+03 | 1.69E+04 | 2.19E+03 | 1.84E+06 | 3.25E+05 | 1.80 | 0.92 | |||
| ZM1 | 1.77E+03 | 5.02E+02 | ND | ND | 5.33E+05 | 3.61E+04 | 0.33 | ND | |||
| ZM2 | 2.00E+03 | 2.28E+02 | ND | ND | 5.17E+05 | 6.79E+04 | 0.39 | ND | |||
| BDW | 7.88E+05 | 2.05E+04 | 9.76E+04 | 0.00E+00 | 6.68E+06 | 2.83E+04 | 11.80 | 1.46 | |||
| QS | ND | ND | 2.00E+04 | 5.40E+03 | 2.35E+06 | 3.82E+05 | ND | 0.85 | |||
| GN | ND | ND | 4.38E+03 | 1.63E+02 | 1.95E+05 | 1.02E+05 | ND | 2.24 | |||
Table 5 Abundance and proportion of total bacterial 16S rRNA, soxB and dsrA genes as determined by qPCR
| 样点 名称 | soxB基因拷贝数/ (copies·mL-1) | dsrA基因拷贝数/ (copies·mL-1) | 16S rRNA基因拷贝数/ (copies·mL-1) | soxB基因相对丰度/ % | dsrA基因相对丰度/ % | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| soxB | SD | dsrA | SD | 16S2) | SD | soxB/16S | dsrA/16S | ||||
| DB1 | 4.53E+04 | 1.16E+04 | 2.24E+05 | 2.83E+03 | 7.41E+05 | 4.22E+05 | 6.12 | 30.25 | |||
| DB41 | 3.62E+03 | 7.85E+02 | ND | ND | 2.31E+05 | 4.88E+04 | 1.57 | ND | |||
| DB45 | ND1) | ND | 2.18E+02 | 1.48E+02 | 9.69E+02 | 6.36E+00 | ND | 22.50 | |||
| DB5 | 2.14E+05 | 3.61E+03 | 8.42E+04 | 3.99E+03 | 4.66E+06 | 2.51E+05 | 4.59 | 1.80 | |||
| DB6 | 3.72E+03 | 1.12E+03 | 2.07E+04 | 1.59E+03 | 2.50E+05 | 7.35E+04 | 1.49 | 8.28 | |||
| DB7 | 1.37E+05 | 3.17E+04 | 6.55E+04 | 7.06E+03 | 3.43E+06 | 4.24E+04 | 4.00 | 1.91 | |||
| DB8 | 4.89E+04 | 1.07E+04 | 1.36E+04 | 1.46E+03 | 2.23E+06 | 1.06E+05 | 2.19 | 0.61 | |||
| DB10 | 3.31E+04 | 2.25E+03 | 1.69E+04 | 2.19E+03 | 1.84E+06 | 3.25E+05 | 1.80 | 0.92 | |||
| ZM1 | 1.77E+03 | 5.02E+02 | ND | ND | 5.33E+05 | 3.61E+04 | 0.33 | ND | |||
| ZM2 | 2.00E+03 | 2.28E+02 | ND | ND | 5.17E+05 | 6.79E+04 | 0.39 | ND | |||
| BDW | 7.88E+05 | 2.05E+04 | 9.76E+04 | 0.00E+00 | 6.68E+06 | 2.83E+04 | 11.80 | 1.46 | |||
| QS | ND | ND | 2.00E+04 | 5.40E+03 | 2.35E+06 | 3.82E+05 | ND | 0.85 | |||
| GN | ND | ND | 4.38E+03 | 1.63E+02 | 1.95E+05 | 1.02E+05 | ND | 2.24 | |||
| [1] |
ABUSAM A, AL-SALMAIN F, MYDLARCZYK A, 2019. Seasonal variations of the growth of filamentous bacteria in Kuwait’s wastewater treatment plants[J]. Desalination and Water Treatment, 176: 370-374.
DOI URL |
| [2] |
ALONSO-SÁEZ L, MORÁN X A G, GONZÁLEZ J M, 2020. Transcriptional patterns of biogeochemically relevant marker genes by temperate marine bacteria[J]. Frontiers in Microbiology, 11: 465.
DOI URL |
| [3] |
CASTELÁN-SÁNCHEZ H G, MEZA-RODRIGUEZ P M, CARRILLO E, et al., 2020. The microbial composition in circumneutral thermal springs from Chignahuapan, Puebla, Mexico reveals the presence of particular sulfur-oxidizing bacterial and viral communities[J]. Microorganisms, 8(11): 1677.
DOI URL |
| [4] |
CHEN Y, MI T Z, LIU Y T, et al., 2020. Microbial community composition and function in sediments from the Pearl River Mouth Basin[J]. Journal of Ocean University of China, 19(4): 941-953.
DOI |
| [5] | CHERNITSYNA S M, KHALZOV I A, POGODAEVA T V, et al., 2020. Distribution and morphology of colorless sulfur bacterium of the genus Thiothrix in water reservoirs of Baikal Rift Zone[J]. Limnology and Freshwater Biology, 4(4): 976-978. |
| [6] | GAO S X, HE Q L, WANG H Y, 2019. Research on the aerobic granular sludge under alkalinity in sequencing batch reactors: removal efficiency, metagenomic and key microbes[J]. Bioresource Technology, 296: 122280. |
| [7] | GUO L, WANG G C, SHENG Y Z, et al., 2021. Hydrogeochemical constraints shape hot spring microbial community compositions: evidence from acidic, moderate-temperature springs and alkaline, high-temperature springs, south-western Yunnan geothermal areas, China[J]. Journal of Geophysical Research: Biogeosciences, 126(3): e2020JG005868-1-e2020JG005868-19. |
| [8] |
GUO Q H, PLANER-FRIEDRICH B P, LIU M L, et al., 2019. Magmatic fluid input explaining the geochemical anomaly of very high arsenic in some southern Tibetan geothermal waters[J]. Chemical Geology, 513: 32-43.
DOI URL |
| [9] |
GUPTA S, PLUGGE C M, KLOK J B M, et al., 2022. Comparative analysis of microbial communities from different full-scale haloalkaline biodesulfurization systems[J]. Applied Microbiology and Biotechnology, 106(4): 1759-1776.
DOI PMID |
| [10] | HOU W G, WANG S, DONG H L, et al., 2013. A comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan province China using 16S rRNA gene pyrosequencing[J]. PloS One, 8(1): 1-15. |
| [11] |
JAFFER Y D, PURUSHOTHAMAN C S, KUMAR H S, et al., 2019. A combined approach of 16S rRNA and a functional marker gene, soxB to reveal the diversity of sulphur-oxidising bacteria in thermal springs[J]. Archives of Microbiology, 201(5) :951-967.
DOI |
| [12] | JIANG Z, LI P, JIANG D W, et al., 2016. Microbial community structure and arsenic biogeochemistry in an acid vapor-formed spring in Tengchong geothermal area, China[J]. PloS One, 11(1): 1-16. |
| [13] | KOJIMA H, WATANABE T, IWATA T, et al., 2014. Identification of major planktonic sulfur oxidizers in stratified freshwater lake[J]. PLoS One, 9(4): e93877. |
| [14] |
KUBO K, KNITTEL K, AMANN R, et al., 2011. Sulfur-metabolizing bacterial populations in microbial mats of the Nakabusa hot spring, Japan[J]. Systematic and Applied Microbiology, 34(4): 293-302.
DOI PMID |
| [15] |
LI P, JIANG Z, WANG Y H, et al., 2017. Analysis of the functional gene structure and metabolic potential of microbial community in high arsenic groundwater[J]. Water Research, 123: 268-276.
DOI PMID |
| [16] | LU G S, LAROWE D E, AMEND J P, 2021. Bioenergetic potentials in terrestrial, shallow-sea and deep-sea hydrothermal systems[J]. Chemical Geology, 583: 120449. |
| [17] |
LUO J F, TAN X Q, LIU K X, et al., 2018. Survey of sulfur-oxidizing bacterial community in the Pearl River water using soxB, sqr, and dsrA as Molecular Biomarkers[J]. 3 Biotech, 8(1): 1-12.
DOI URL |
| [18] | MA L, WU G, YANG J, et al., 2021. Distribution of hydrogen-producing bacteria in Tibetan hot springs, China[J]. Frontiers in Microbiology, 12: 1-12. |
| [19] |
PANDEY S K, NARAYAN K D, BANDYOPADHYAY S, et al., 2009. Thiosulfate oxidation by Comamonas sp s23 isolated from a sulfur spring[J]. Current Microbiology, 58(5): 516-521.
DOI URL |
| [20] | QING C, NICOL A, LI P, et al., 2023. Different sulfide to arsenic ratios driving arsenic speciation and microbial community interactions in two alkaline hot springs[J]. Environmental Research, 218: 115033. |
| [21] |
SHU W S, HUANG L N, 2022. Microbial diversity in extreme environments[J]. Nature Reviews Microbiology, 20(4): 219-235.
DOI |
| [22] |
SKIRNISDOTTIR S, HREGGVIDSSON G O, HJÖRLEIFSDOTTIR S, et al., 2000. Influence of sulfide and temperature on species composition and community structure of hot spring microbial mats[J]. Applied and Environmental Microbiology, 66(7): 2835-2841.
DOI PMID |
| [23] |
SOROKIN D Y, TOUROVA T P, KUZNETSOV B B, et al., 2000. Roseinatronobacter thiooxidans gen. nov., sp, nov., a new alkaliphilic aerobic bacteriochlorophylla-containing bacterium isolated from a soda lake[J]. Microbiology, 69(1): 75-82.
DOI URL |
| [24] | TAKEDA M, KAMAGATA Y, GHIROSE W C, et al., 2002. Caldimonas manganoxidans gen. nov., sp nov., a poly(3-hydroxybutyrate)-degrading, manganese-oxidizing thermophile[J]. International Journal of Systematic and Evolutionary Microbiology, 52(3): 895-900. |
| [25] |
TOUROVA T P, SLOBODOVA N V, BUMAZHKIN B K., et al., 2013. Analysis of community composition of sulfur-oxidizing bacteria in hypersaline and soda lakes using soxB as a functional molecular marker[J]. FEMS Microbiology Ecology, 84(2): 280-289.
DOI URL |
| [26] |
VIOLLIER E, INGLETT P W, HUNTER K, et al., 2000. The ferrozine method revisited: Fe(II)/Fe(III) determination in natural waters[J]. Applied geochemistry, 15(6): 785-790.
DOI URL |
| [27] |
WANG L, YU M, LIU Y, et al., 2018. Comparative analyses of the bacterial community of hydrothermal deposits and seafloor sediments across Okinawa Trough[J]. Journal of Marine Systems, 180: 162-172.
DOI URL |
| [28] |
WANG R, LIN J Q, LIU X M, et al., 2019. Sulfur oxidation in the acidophilic autotrophic Acidithiobacillus spp[J]. Frontiers in Microbiology, 9: 3290.
DOI URL |
| [29] |
WANG Y H, LI P, JIANG Z, et al., 2018. Diversity and abundance of arsenic methylating microorganisms in high arsenic groundwater from Hetao Plain of Inner Mongolia, China[J]. Ecotoxicology, 27(8): 1047-1057.
DOI PMID |
| [30] |
WANG Y X., LI P, GUO Q H., et al., 2018. Environmental biogeochemistry of high arsenic geothermal fluids[J]. Applied Geochemistry, 97: 81-92.
DOI URL |
| [31] |
WATANABE T, KOJIMA H, SHINOHARA A, et al., 2016. Sulfurirhabdus autotrophica gen. nov., sp nov., isolated from a freshwater lake[J]. International Journal of Systematic and Evolutionary Microbiology, 66(1): 113-117.
DOI URL |
| [32] | WU B, LIU F F, FANG W W, et al., 2021. Microbial sulfur metabolism and environmental implications[J]. Science of The Total Environment, 778(12-13): 146085. |
| [33] |
YANG J, JIANG H C, DONG H L, et al., 2013. Abundance and diversity of sulfur-oxidizing bacteria along a salinity gradient in four Qinghai-Tibetan lakes, China[J]. Geomicrobiology Journal, 30(9): 851-860.
DOI URL |
| [34] |
ZECCHIN S, COLOMBO M, CAVALCA L, 2019. Exposure to different arsenic species drives the establishment of iron and sulfur-oxidizing bacteria on rice root iron plaques[J]. World Journal of Microbiology and Biotechnology, 35(8): 1-12.
DOI |
| [35] |
ZHANG D R, XIA J L, NIE ZH Y, et al., 2019. Mechanism by which ferric iron promotes the bioleaching of arsenopyrite by the moderate thermophile Sulfobacillus Thermosulfidooxidans [J]. Process Biochem, 81: 11-21.
DOI URL |
| [36] | ZHAO Z Q, SUN C, LI Y, et al., 2020. Driving microbial sulfur cycle for phenol degradation coupled with Cr(VI) reduction via Fe(III)/Fe(II) transformation[J]. Chemical Engineering Journal, 393: 124801. |
| [37] | ZHOU L, WANG D W, ZHANG S L, et al., 2020. Functional microorganisms involved in the sulfur and nitrogen metabolism in production water from a high-temperature offshore petroleum reservoir[J]. International Biodeterioration & Biodegradation, 154: 105057. |
| [38] | 刘阳, 姜丽晶, 邵宗泽, 2018. 硫氧化细菌的种类及硫氧化途径的研究进展[J]. 微生物学报, 58(2): 191-201. |
| LIU Y, JIANG L J, SHAO Z Z, 2018. Advances in sulfur-oxidizing bacterial taxa and their sulfur oxidation pathways[J]. Acta Microbiologica Sinica, 58(2): 191-201. | |
| [39] | 吕苑苑, 郑绵平, 赵平, 等, 2014. 滇藏地热带地热水硼同位素地球化学过程及其物源示踪[J]. 中国科学: 地球科学, 44(9): 1968-1979. |
| LÜ Y Y, ZHENG M P, ZHAO P, et al., 2014. Geochemical processes and origin of boron isotopes in geothermal water in the Yunnan-Tibet geothermal zone[J]. Science China: Earth Sciences, 44(9): 1968-1979. | |
| [40] | 韩天赐, 2018. 西藏曲才热泉的细菌多样性及栖热菌属比较基因组学[D]. 昆明: 云南大学. |
| HAN T C, 2018. Bacterial diversity of Qucai hot springs in Tibet & comparative genomics of the genus Thermus[D]. Kunming: Yunnan University. | |
| [41] | 姜舟, 2016. 腾冲地热区砷的环境生物地球化学研究[D]. 武汉: 中国地质大学(武汉). |
| JIANG Z, 2016. Environmental biogeochemistry of arsenic in Tengchong geothermal area, china[D] Wuhan: China University of Geosciences (Wuhan). | |
| [42] | 邵博, 2019. 微氧强化自养-异养联合反硝化处理含不同硫氮比废水的效能研究[D]. 哈尔滨: 哈尔滨工业大学. |
| SHAO B, 2019. Research on mitigating adverseimpact of varying sulfide to nitrateratio on the integrated autotrophicand heterotrophic denitrification process via microaerobic condition[D]. Harbin: Harbin Institute of Technology. | |
| [43] | 王尚, 2015. 滇藏热泉微生物群落分布及其控制因素研究[D]. 北京: 中国地质大学 (北京). |
| WANG S, 2015. The distribution patterns and environmental constraints of microbial community in hot springs in Western China[D]. Beijing: China University of Geosciences (Beijing). | |
| [44] | 杨磊, 张爽, 刘涛, 等, 2020. 五大连池东焦得布山硫氧化细菌多样性及其与环境因子的关系[J]. 黑龙江八一农垦大学学报, 32(5): 77-82. |
| YANG L, ZHANG S, LIU T, et al., 2020. Sulfur-oxidizing bacteria diversity in Dongjiao-debu Volcanoes located in Wudalianchi[J]. Journal of Heilongjiang Bayi Agricultural University, 32(5): 77-82. | |
| [45] | 张玉, 米铁柱, 甄毓, 等, 2018. 海洋沉积物中硫酸盐还原菌和硫氧化菌群落分析方法的比较[J]. 环境科学, 39(1): 438-449. |
| ZHANG Y, MI T Z, ZHEN Y, et al., 2018. Analysis of sulfate-reducing and sulfur-oxidizing prokaryote community structures in marine sediments with different sequencing technologies[J]. Environmental Science, 39(1): 438-449. | |
| [46] | 甄莉, 吴耿, 杨渐, 等, 2019. 西藏热泉沉积物的硫氧化细菌多样性及其影响因素[J]. 微生物学报, 59(6): 1089-1104. |
| ZHEN L, WU G, YANG J, et al., 2019. Distribution and diversity of sulfur-oxidizing bacteriain the surface sediments of Tibetan hot springs[J]. Acta Microbiologica Sinica, 59(6): 1089-1104. |
| [1] | HOU Hui, YAN Peixuan, XIE Qinmi, ZHAO Hongliang, PANG Danbo, CHEN Lin, LI Xuebin, HU Yang, LIANG Yongliang, NI Xilu. Characterization of Arbuscular Mycorrhizal Fungal Community Diversity in the Rhizosphere Soils of Prunus mongolica Scrub of Helan Mountain [J]. Ecology and Environment, 2023, 32(5): 857-865. |
| [2] | JIANG Yongwei, DING Zhenjun, YUAN Junbin, ZHANG Zheng, LI Yang, WEN Qingchun, WANG Yeyao, JIN Xiaowei. Study on Benthic Macroinvertebrates Community Structure and Water Quality Evaluation in Main Rivers of Liaoning Province [J]. Ecology and Environment, 2023, 32(5): 969-979. |
| [3] | WANG Yun, ZHENG Xilai, CAO Min, LI Lei, SONG Xiaoran, LIN Xiaolei, GUO Kai. Study on Denitrification Performance and Control Factors in Brackish-Freshwater Transition Zone of Coastal Aquifer [J]. Ecology and Environment, 2023, 32(5): 980-988. |
| [4] | WANG Xinyu, GAO Dengzhou, LIU Bolin, WANG Bin, ZHENG Yanling, LI Xiaofei, HOU Lijun. The Tidal-cycle Variation and Influencing Factors of Dark Carbon Fixation Process in the Yangtze Estuary [J]. Ecology and Environment, 2023, 32(4): 733-743. |
| [5] | HU Fang, LIU Jutao, WEN Chunyun, HAN Liu, WEN Hui. Phytoplankton Community Structure and Evaluation of Aquatic Ecological Conditions in Fu River Basin [J]. Ecology and Environment, 2023, 32(4): 744-755. |
| [6] | YU Fei, ZENG Hailong, FANG Huaiyang, FU Lingfang, LIN Shu, DONG Jiahao. Spatio-temporal Variation Characteristics of Phytoplankton Functional Groups and Water Quality Evaluation in the Typical Tidal River Network [J]. Ecology and Environment, 2023, 32(4): 756-765. |
| [7] | WANG Lixiao, LIU Jinxian, CHAI Baofeng. Response of Soil Bacterial Community and Nitrogen Cycle during the Natural Recovery of Abandoned Farmland in Subalpine of the North China [J]. Ecology and Environment, 2022, 31(8): 1537-1546. |
| [8] | JIANG Nihao, ZHANG Shihao, ZHANG Shihan. Interspecific Associations and Environmental Interpretation of the Dominant Species of the Communities Invaded by Ageratina adenophora in Ailao Mountains [J]. Ecology and Environment, 2022, 31(7): 1370-1382. |
| [9] | YANG Chong, WANG Chunyan, WANG Wenying, MAO Xufeng, ZHOU Huakun, CHEN Zhe, SUONANJi , JIN Lei, MA Huaqing. Soil Nutrient Characteristics and Quality Evaluation of Alpine Grassland in the Source Area of the Yellow River on the Qinghai Tibet Plateau [J]. Ecology and Environment, 2022, 31(5): 896-908. |
| [10] | WANG Yingcheng, YAO Shiting, JIN Xin, YU Wenzhen, LU Guangxin, WANG Junbang. Comparative Study on Soil Bacterial Diversity of Degraded Alpine Meadow in the Sanjiangyuan Region [J]. Ecology and Environment, 2022, 31(4): 695-703. |
| [11] | LIU Hongmei, HAI Xiang, AN Kerui, ZHANG Haifang, WANH Hui, ZHANG Yanjun, WANG Lili, ZHANG Guilong, YANG Dianlin. Effects of Different Fertilization Regimes on Community Structure Diversity of CO2-assimilating Bacteria in Maize Field of Fluvo-aquic Soil in North China [J]. Ecology and Environment, 2022, 31(4): 715-722. |
| [12] | XIA Kai, DENG Pengfei, MA Ruihao, WANG Fei, WEN Zhengyu, XU Xiaoniu. Changes of Soil Bacterial Community Structure and Diversity from Conversion of Masson Pine Secondary Forest to Slash Pine and Chinese Fir Plantations [J]. Ecology and Environment, 2022, 31(3): 460-469. |
| [13] | SONG Xiuli, HUANG Ruilong, KE Caijie, HUANG Wei, ZHANG Wu, TAO Bo. Effects of Different Cropping Systems on Bacterial Community Structure and Diversity in Continuous Cropping Soil [J]. Ecology and Environment, 2022, 31(3): 487-496. |
| [14] | XUE Wenkai, ZHU Pan, DE Ji, GUO Xiaofang. Study on the Temporal and Spatial Characteristics of the Dominant Species of Cultivable Filamentous Fungi in Nam Co Lake [J]. Ecology and Environment, 2022, 31(12): 2331-2340. |
| [15] | LI Cong, LÜ Jinghua, LU Mei, YANG Zhidong, LIU Pan, REN Yulian, DU Fan. Responses of Soil Bacterial Communities to Vertical Vegetarian Zone Changes in the Subtropical Forests, Southeastern Yunnan [J]. Ecology and Environment, 2022, 31(10): 1971-1983. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2021 Editorial Office of ACTA PETROLEI SINICA
Address:No. 6 Liupukang Street, Xicheng District, Beijing, P.R.China, 510650
Tel: 86-010-62067128, 86-010-62067137, 86-010-62067139
Fax: 86-10-62067130
Email: syxb@cnpc.com.cn
Support byBeijing Magtech Co.ltd, E-mail:support@magtech.com.cn
