Ecology and Environment ›› 2023, Vol. 32 ›› Issue (5): 866-877.DOI: 10.16258/j.cnki.1674-5906.2023.05.005
• Research Articles • Previous Articles Next Articles
ZHAO Hongbin1,2,3,*( ), BAI Xue1, FAN Yufeng1, ZHANG Xiaofu1, ZHANG Tao1, LI Shufen1,3
), BAI Xue1, FAN Yufeng1, ZHANG Xiaofu1, ZHANG Tao1, LI Shufen1,3
Received:2022-12-13
Online:2023-05-18
Published:2023-08-09
Contact:
ZHAO Hongbin
赵鸿彬1,2,3,*( ), 白雪1, 范宇凤1, 张晓馥1, 张涛1, 李淑芬1,3
), 白雪1, 范宇凤1, 张晓馥1, 张涛1, 李淑芬1,3
通讯作者:
赵鸿彬
作者简介:赵鸿彬(1978年生),女,副教授,研究方向为草原生态与牧草分子育种。E-mail: hbzhao04@163.com
基金资助:CLC Number:
ZHAO Hongbin, BAI Xue, FAN Yufeng, ZHANG Xiaofu, ZHANG Tao, LI Shufen. Cloning and Differential Expression Analysis of Stipa Breviflora StbCRY1 and StbCRY2 Genes[J]. Ecology and Environment, 2023, 32(5): 866-877.
赵鸿彬, 白雪, 范宇凤, 张晓馥, 张涛, 李淑芬. 短花针茅StbCRY1和StbCRY2基因克隆与表达差异分析[J]. 生态环境学报, 2023, 32(5): 866-877.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.jeesci.com/EN/10.16258/j.cnki.1674-5906.2023.05.005
| 序列名称 | 序列用途 | 序列 |
|---|---|---|
| StbCRY1-F1 | ORF cloning | CACTGCCCACGCACTCA |
| StbCRY1-R1 | ORF cloning | GCAACAACCAAAGGCAAT |
| StbCRY2-F1 | ORF cloning | GTTGTTCCATTCCGTCAC |
| StbCRY2-R1 | ORF cloning | TGATGAGAGTGGAATACGGGA |
| 5′ StbCRY1-GSP1 | 5′ cDNA RACE | GTTTCAGCCCTGCCTAAAGAG |
| 5′ StbCRY1-GSP2 | 5′ cDNA RACE | CCTTCTTCCATACCGTTCTCCAT |
| 5′ StbCRY2-GSP1 | 5′ cDNA RACE | CATCCCGCACAAGTGAAATAG |
| 5′ StbCRY2-GSP1 | 5′ cDNA RACE | AGCGACTTCTTGAGCCACCAC |
| StbCRY1-GFP-F | Gene cloning | TTCATTTGGAGAA CACGGGGGAC |
| StbCRY1-GFP-R | Gene cloning | CAAGACCGGCAA CAGGATTCAATA |
| StbCRY2-GFP-F | Gene cloning | TTCATTTGGAGAA CACGGGGGAC |
| StbCRY2-GFP-R | Gene cloning | CAAGACCGGCAA CAGGATTCAATA |
| internal TLF Upstream | Gene expression analysis | CCCTCAGTGTG TGTTTGACC |
| internal TLF Downstream | Gene expression analysis | CTTGAGACCC TTCCTCTTGC |
| StbCRY1 Upstream | Gene expression analysis | TCATCAGCAGC GTAAGCATCT |
| StbCRY1 Downstream | Gene expression analysis | CGTCCACCAC CAGCAGTAT |
| StbCRY2 Upstream | Gene expression analysis | GCCAGTCCCAGG TATAGAGAATC |
| StbCRY2 Downstream | Gene expression analysis | AGGAGACCAC GAAGCACAA |
Table 1 The sequence and usages of the primers
| 序列名称 | 序列用途 | 序列 |
|---|---|---|
| StbCRY1-F1 | ORF cloning | CACTGCCCACGCACTCA |
| StbCRY1-R1 | ORF cloning | GCAACAACCAAAGGCAAT |
| StbCRY2-F1 | ORF cloning | GTTGTTCCATTCCGTCAC |
| StbCRY2-R1 | ORF cloning | TGATGAGAGTGGAATACGGGA |
| 5′ StbCRY1-GSP1 | 5′ cDNA RACE | GTTTCAGCCCTGCCTAAAGAG |
| 5′ StbCRY1-GSP2 | 5′ cDNA RACE | CCTTCTTCCATACCGTTCTCCAT |
| 5′ StbCRY2-GSP1 | 5′ cDNA RACE | CATCCCGCACAAGTGAAATAG |
| 5′ StbCRY2-GSP1 | 5′ cDNA RACE | AGCGACTTCTTGAGCCACCAC |
| StbCRY1-GFP-F | Gene cloning | TTCATTTGGAGAA CACGGGGGAC |
| StbCRY1-GFP-R | Gene cloning | CAAGACCGGCAA CAGGATTCAATA |
| StbCRY2-GFP-F | Gene cloning | TTCATTTGGAGAA CACGGGGGAC |
| StbCRY2-GFP-R | Gene cloning | CAAGACCGGCAA CAGGATTCAATA |
| internal TLF Upstream | Gene expression analysis | CCCTCAGTGTG TGTTTGACC |
| internal TLF Downstream | Gene expression analysis | CTTGAGACCC TTCCTCTTGC |
| StbCRY1 Upstream | Gene expression analysis | TCATCAGCAGC GTAAGCATCT |
| StbCRY1 Downstream | Gene expression analysis | CGTCCACCAC CAGCAGTAT |
| StbCRY2 Upstream | Gene expression analysis | GCCAGTCCCAGG TATAGAGAATC |
| StbCRY2 Downstream | Gene expression analysis | AGGAGACCAC GAAGCACAA |
| 一级结构特性 | ORF长度/bp | 推导氨基酸数 | 理论等电点 (PI) | 分子量 | 不稳定系数 (II) | 总平均亲水性 | 脂肪系数 (AI) | 负电荷残基 | 正电荷残基 |
|---|---|---|---|---|---|---|---|---|---|
| CRY1 | 2102 | 699 | 5.48 | 78829.36 | 51.84 | -0.464 | 76.07 | 93 | 72 |
| CRY2 | 1962 | 653 | 5.53 | 73429.95 | 45.5 | -0.387 | 82.25 | 88 | 72 |
Table 2 Structural characteristics of proteins encoded by StbCRY1 and StbCRY2 genes
| 一级结构特性 | ORF长度/bp | 推导氨基酸数 | 理论等电点 (PI) | 分子量 | 不稳定系数 (II) | 总平均亲水性 | 脂肪系数 (AI) | 负电荷残基 | 正电荷残基 |
|---|---|---|---|---|---|---|---|---|---|
| CRY1 | 2102 | 699 | 5.48 | 78829.36 | 51.84 | -0.464 | 76.07 | 93 | 72 |
| CRY2 | 1962 | 653 | 5.53 | 73429.95 | 45.5 | -0.387 | 82.25 | 88 | 72 |
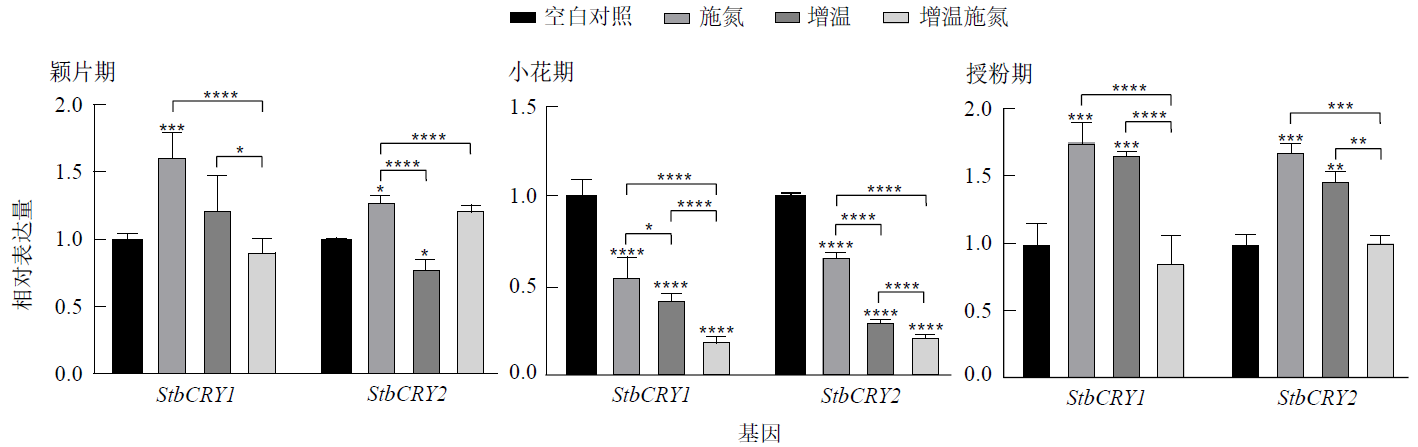
Figure 7 Expression differences of StbCRY1 and StbCRY2 in reproductive branches at the same flower development stage under different temperature and nitrogen application conditions
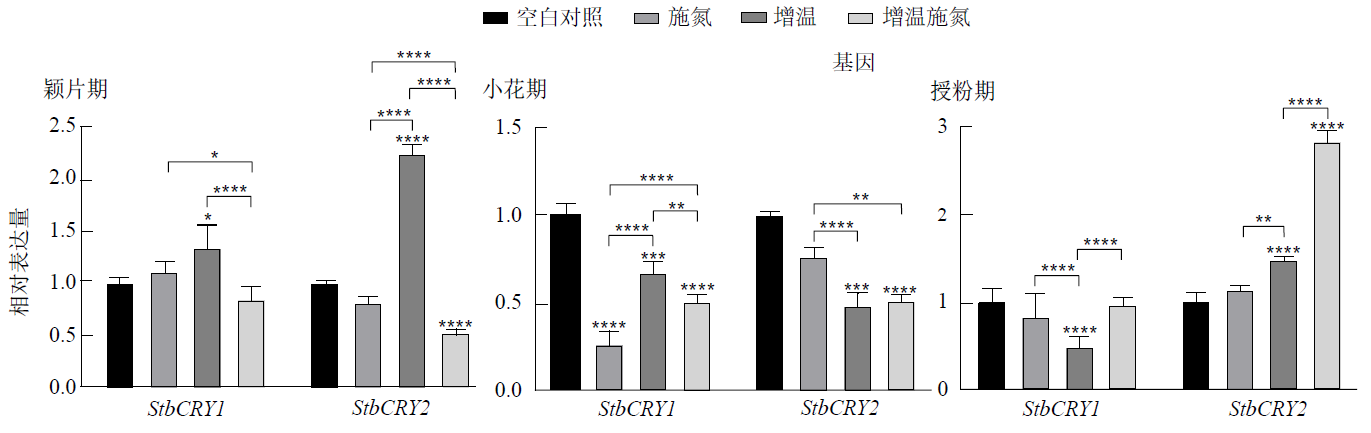
Figure 8 Expression differences of StbCRY1 and StbCRY2 in vegetative branch leaves leaves at the same flower development stage under different temperature and nitrogen application conditions
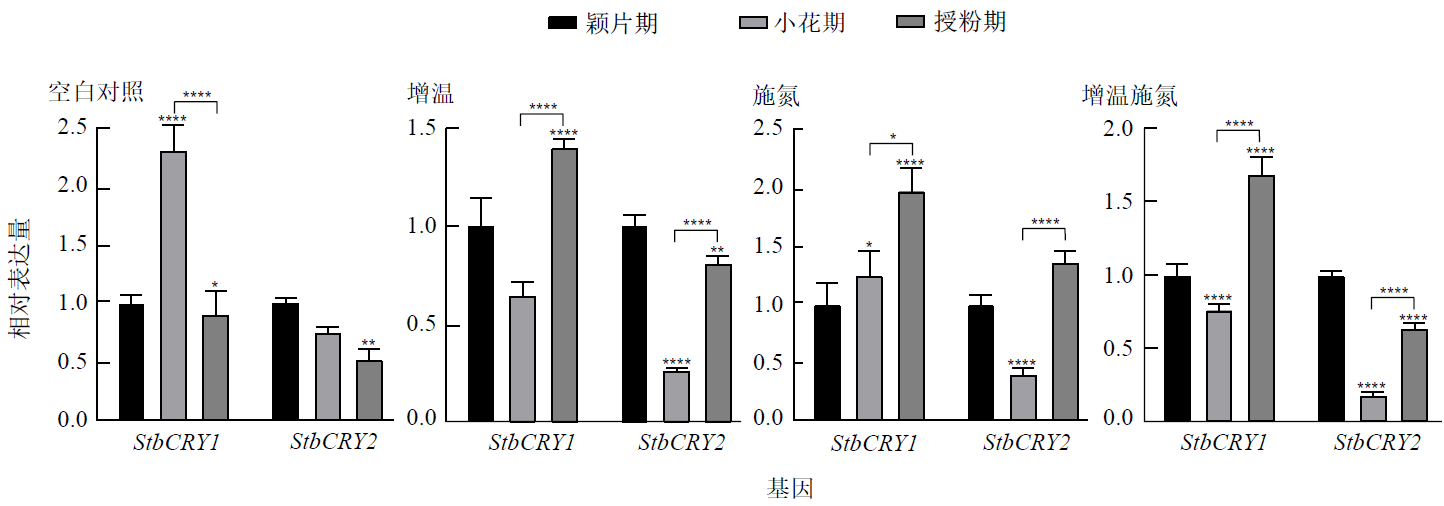
Figure 9 Expression differences of StbCRY1 and StbCRY2 in reproductive branches at different flower development stages under the same temperature and nitrogen application conditions
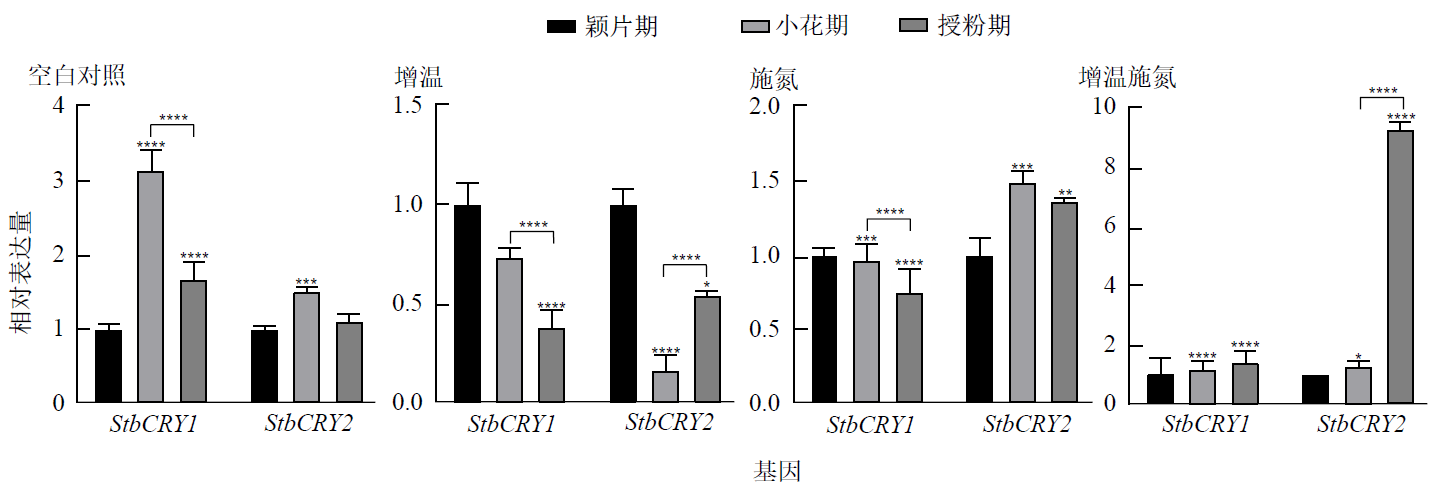
Figure 10 Expression differences of StbCRY1 and StbCRY2 in vegetative branch leaves leaves at different flower development stages under the same temperature and nitrogen application conditions
| [1] |
CASHMORE A R, JARILLO J A, WU Y J, et al., 1999. Cryptochromes: blue light receptors for plants and animal[J]. Science, 284(5415): 760-7653.
DOI URL |
| [2] |
GALLOWAY J N, ABER J D, ERISMAN J W, et al., 2003. The nitrogen cascade[J]. Bioscience, 53(4): 341-356.
DOI URL |
| [3] |
LIN C, AHMAD M, CASHMORE A R, et al., 1996. Cashmore AR. Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development[J]. The Plant Journal, 10(5): 893-902.
DOI URL |
| [4] | LIN C, AHMAD M, GORDON D, et al., 1995. Expression of an Arabidopsis cryptochrome gene in transgenic tobacco results in hypersensitivity to blue UV-A, and green light. Proceedings of the National[J]. Academy of Sciences, 92(18): 8423-8427. |
| [5] |
LIU B B, YANG Z H, GOMEZ A, et al., 2016. Signaling mechanisms of plant cryptochromes in Arabidopsis thaliana [J]. Journal of Plant Research, 129(2): 137-148.
DOI URL |
| [6] |
LIU B, ZUO Z C, LIU H T, et al., 2011. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light[J]. Genes & Development, 25(10): 1029-1034.
DOI URL |
| [7] | LIU Y W, LI X, LI K W, et al., 2013. Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis[J]. PLOS Genetics, 9(10): e1003861. |
| [8] |
LIVAK K J, SCHMITTGEN T D, 2001. Analysis of relative gene expression data using Real Time Quantitative PCR and the 2-ΔΔCT method[J]. Methods, 25(4): 402-408.
DOI URL |
| [9] |
MA L B, LI X, ZHAO Z W, et al., 2021. Light-Response Bric-A-Brack/Tramtrack/Broad proteins mediate cryptochrome 2 degradation in response to low ambient temperature[J]. The Plant Cell, 33(12): 3610-3620.
DOI PMID |
| [10] |
JOHANSSON M, STAIGER D, 2015. Time to flower: interplay between photoperiod and the circadian clock[J]. Journal of Experimental Botany, 66(3): 719-730.
DOI PMID |
| [11] |
SHERRY R A, ZHOU X, GU S, et al., 2007. Divergence of reproductive phenology under climate warming[J]. Proceedings of the National Academy of Sciences of the United States of America, 104(1): 198-202.
DOI PMID |
| [12] |
SMITH H, 2000. Phytochromes and light signal perception by plants-an emerging synthesis[J]. Nature, 407(6804): 585-591.
DOI |
| [13] |
WANG H Y, MA L G, LI J M, et al., 2001. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development[J]. Science, 294(5540): 154-158.
DOI URL |
| [14] |
YAN S, RUI Z, SHAN G, et al., 2022. Transcriptome analysis and phenotyping of walnut seedling roots under nitrogen stresses[J]. Scientific reports, 12(1): 12066.
DOI PMID |
| [15] |
YANG H Q, WU Y J, TANG H R, et al., 2000. The C termini of Arabidopsis cryptochromes mediate a constitutive light response[J]. Cell, 103(5): 815-827.
DOI URL |
| [16] | YU X H, LIU HT, KLEJNOT J, et al., 2010. The cryptochrome blue light receptors[J]. The Arabidopsis Book, 8: e0135. |
| [17] |
ZLATANOVA J, THAKAR A, 2008. H2A.Z: view from the top[J]. Structure, 16(2): 166-179.
DOI PMID |
| [18] |
ZUO Z C, LIU H T, LIU B, et al., 2011. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis[J]. Current Biology, 21(10): 841-847.
DOI URL |
| [19] | 白春利, 阿拉塔, 陈海军, 等, 2013. 氮素和水分添加对短花针茅荒漠草原植物群落特征的影响[J]. 中国草地学报, 35(2): 69-75. |
| BAI C L, ALATA, CHEN H J, et al., 2013. Effect of addition of nitrogen and water on plant community characteristics of Stipa breviflora desert steppe[J]. Chinese Journal of Grassland, 35(2): 69-75. | |
| [20] | 白雪, 2022. 短花针茅StbCRY1和StbCRY2基因的克隆与表达差异的初步分析[D]. 呼和浩特: 内蒙古农业大学:20-23. |
| BAI X, 2022. Clonging of StbCRY1and StbCRY2 genes of Stipa breviflora and analysis of expression differences[D]. Hohhot: Inner Mongolia Agricultural University:20-23. | |
| [21] | 黄琛, 2015. 不同载畜率对短花针茅荒漠草原植被空间异质性影响的研究[D]. 呼和浩特: 内蒙古农业大学: 3-7. |
| HUANG C, 2015. Study on Vegetatiion Spatial heterogeneity under different stocking rate in Stipa breviflora desert steppe[D]. Hohhot: Inner Mongolia Agricultural University: 3-7. | |
| [22] | 蒋玮琳, 龙鸿, 2014. 隐花色素基因CRY2延迟拟南芥营养生长时相转变[J]. 热带作物学报, 35(11): 2211-2214. |
| JIANG W L, LONG H, 2014. Regulation of vegetative phase change by CRY2 in Atabidopsis thaliana[J]. Chinese Journal of Tropical Crops, 35(11): 2211-2214. | |
| [23] | 李仕铭, 周增, 涂敏, 等, 2018. 拟南芥隐花色素CRY光信号通路的研究进展[J]. 分子植物育种, 16(13): 4444-4452. |
| LI S M, ZHOU Z, TU M, et al., 2018. Research progress of photosignal pathway of cryptochrome in Arabidopsis thaliana[J]. Molecular Plant Breeding, 16(13): 4444-4452. | |
| [24] | 李元恒, 韩国栋, 王珍, 等, 2014. 增温和氮素添加对内蒙古荒漠草原植物生殖物候的影响[J]. 生态学杂志, 33(4): 849-856. |
| LI Y H, HAN G D, WANG Z, et al., 2014. Influences of warming and nitrogen addition on plant reproductive phenology in Inner Mongolia desert steppe[J]. Chinese Journal of Ecology, 33(4): 849-856. | |
| [25] | 刘转霞, 余运康, 陈裕坤, 等, 2017. 福州野生蕉隐花色素和光敏色素基因家族的克隆及其表达分析[J]. 热带作物学报, 38(11): 2089-2099. |
| LIU Z X, YU Y K, CHEN Y K, et al., 2017. Cloning and expression analysis of Cryptochrome and phytochrome family genes in the wild banana (Musa spp.) from Fuzhou city[J]. Chinese Journal of Tropical Crops, 38(11): 2089-2099. | |
| [26] |
马朝峰, 戴思兰, 2019. 光受体介导信号转导调控植物开花研究进展[J]. 植物学报, 54(1): 9-22.
DOI |
| MA C F, DAI S L, 2019. Advances in Photoreceptor-mediated Signaling Transduction in Flowering Time Regulation[J]. Chinese Bulletin of Botany, 54(1): 9-22. | |
| [27] | 王红霞, 郝建梅, 丁谦, 2017. 植物隐花色素及其分子调控机制[J]. 山东农业科学, 49(3): 148-153. |
| WANG H X, HAO J M, DING Q, 2017. Research progress of cryptochrome in plants and its molecular regulation mechanism[J]. Shandong Agricultural Sciences, 49(3): 148-153. | |
| [28] | 熊璐, 2008. 大豆原生质体瞬时表达体系的建立及大豆隐花色素的亚细胞定位[D]. 长春: 吉林大学:34-42. |
| XIONG L, 2008. Establishment of the protoplasts transient gene expression system and the subcellular localization of cryptochromes in soybean[D]. Changchun: Jilin University:34-42. | |
| [29] | 徐沛, 2008. 小麦隐花色素基因及线粒体ATP合酶Fad亚基基因的克隆与分析[D]. 南京: 南京农业大学:56-59. |
| XU P, 2008. Clonging and molecular characterization of a Fad subunit gene and crypyochrome genes of wheat[D]. Nanjing: Nanjing Agricultural University:56-59. | |
| [30] | 张丽, 吴英英, 刘斌, 2021. 大豆隐花色素CRY2基因表达对马铃薯抗逆性的影响[J]. 分子植物育种, 19(8): 2472-2479. |
| ZHANG L, WU Y Y, LIU B, 2021. Effect of Soybean CRY2 Gene Expression on Stress Resistance of Potato[J]. Molecular Plant Breeding, 19(8): 2472-2479. | |
| [31] | 赵鸿彬, 范宇凤, 张晓馥, 等, 2022. 短花针茅StbCO和StbFT基因全长的克隆与表达差异分析[J]. 中国草地学报, 44(7): 1-12. |
| ZHAO H B, FAN Y F, ZHANG X F, et al., 2022. Cloning and expression analysis of full-length StbCO and StbFT genes in Stipa breviflora[J]. Chinese Journal of Grassland, 44(7): 1-12. | |
| [32] | 朱文琰, 王娅琳, 杨畅, 等, 2021. 不同放牧模式对贵南县高寒草甸优势种羊茅叶属性的影响[J]. 中国草地学报, 43(6): 69-75. |
| ZHU W Y, WANG Y L, YANG C, et al., 2021. Effects of different grazing pattern on leaf characteristics of Festuca ovina in alpine meadow of Guinan County[J]. Chinese Journal of Grassland, 43(6): 69-75. |
| [1] | HOU Hui, YAN Peixuan, XIE Qinmi, ZHAO Hongliang, PANG Danbo, CHEN Lin, LI Xuebin, HU Yang, LIANG Yongliang, NI Xilu. Characterization of Arbuscular Mycorrhizal Fungal Community Diversity in the Rhizosphere Soils of Prunus mongolica Scrub of Helan Mountain [J]. Ecology and Environment, 2023, 32(5): 857-865. |
| [2] | WANG Qi, ZHANG Feng, ZHAO Mengli, ZHANG Xinyu, ZHANG Jun. Effects of Grazing Intensity Community Composition and Inter-species Relationships of Stipa breviflora Desert Steppe, Inner Mongolia, China [J]. Ecology and Environment, 2021, 30(10): 1961-1967. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2021 Editorial Office of ACTA PETROLEI SINICA
Address:No. 6 Liupukang Street, Xicheng District, Beijing, P.R.China, 510650
Tel: 86-010-62067128, 86-010-62067137, 86-010-62067139
Fax: 86-10-62067130
Email: syxb@cnpc.com.cn
Support byBeijing Magtech Co.ltd, E-mail:support@magtech.com.cn
