生态环境学报 ›› 2024, Vol. 33 ›› Issue (10): 1580-1589.DOI: 10.16258/j.cnki.1674-5906.2024.10.010
刘苏杰1,2,3( ), 刘传平3, 方利平3, 陈冠虹3,*(
), 刘传平3, 方利平3, 陈冠虹3,*( ), 李芳柏3
), 李芳柏3
收稿日期:2024-02-15
出版日期:2024-10-18
发布日期:2024-11-15
通讯作者:
*陈冠虹。E-mail: ghchen@soil.gd.cn作者简介:刘苏杰(1998年生),男,硕士研究生,研究方向为土壤砷污染控制。E-mail: liusujie21@mails.ucas.ac.cn
基金资助:
LIU Sujie1,2,3( ), LIU Chuanping3, FANG Liping3, CHEN Guanhong3,*(
), LIU Chuanping3, FANG Liping3, CHEN Guanhong3,*( ), LI Fangbai3
), LI Fangbai3
Received:2024-02-15
Online:2024-10-18
Published:2024-11-15
摘要:
短链脂肪酸互营产甲烷是厌氧有机质分解过程中的关键限速步骤,互营产甲烷菌群砷抗性机制是其适应砷胁迫的重要途径,然而它们的砷转化特征和抗性策略尚不清楚。丁酸是有机质分解产甲烷过程中生成的重要中间产物,丁酸产甲烷菌群的砷甲基化能力及对砷胁迫的耐受机制对于理解厌氧微生物砷转化过程至关重要。利用丁酸为唯一碳源和无机As(III)或有机三价砷MMAs(III)为砷底物分别富集稻田土壤源产甲烷菌群,发现丁酸产甲烷富集物能够将As(III)转化为一甲基砷(MMAs)、二甲基砷(DMAs)和三甲基砷(TMAsO),也能将MMAs(III)转化为DMAs和TMAsO。通过细菌、古菌和功能微生物群落分子解析技术,结果表明共养单胞菌科Syntrophomonadaceae作为丁酸互营氧化功能菌是富集物中主要的细菌类群,甲烷八叠球菌属Methanosarcina和甲烷杆菌属Methanobacterium是主要的产甲烷古菌,它们协同驱动了砷胁迫下的丁酸氧化产甲烷过程。不同砷胁迫条件下的关键微生物类群对比结果显示,MMAs(Ⅲ)胁迫条件进一步富集了Syntrophomonadaceae(55.2%-61.0%)和Methanobacterium(49.0%-55.7%),并且这两种类群中存在潜在砷甲基化微生物,它们可能通过砷甲基化来耐受环境中的砷胁迫。所以,短链脂肪酸互营氧化产甲烷菌群中互营细菌和产甲烷古菌成员可能利用砷甲基化进行解毒,以适应厌氧环境中的高毒性三价无机砷或甲基砷,是其耐受砷胁迫的重要生存策略。研究结果可为理解厌氧微生物类金属耐受机制及其参与的碳砷元素耦合转化过程提供新视角。
中图分类号:
刘苏杰, 刘传平, 方利平, 陈冠虹, 李芳柏. 水稻土丁酸互营产甲烷菌群的砷甲基化特征及机制解析[J]. 生态环境学报, 2024, 33(10): 1580-1589.
LIU Sujie, LIU Chuanping, FANG Liping, CHEN Guanhong, LI Fangbai. Arsenic Methylation Process and the Associated Microbial Mechanisms in Paddy Soil Butyrate-degrading Methanogenic Communities[J]. Ecology and Environment, 2024, 33(10): 1580-1589.
| 组分 | 量值 | 单位 |
|---|---|---|
| 1,4-哌嗪二乙磺酸 (PIPES) | 3.024 | g·L−1 |
| NH4Cl | 0.535 | g·L−1 |
| NaHCO3 | 0.420 | g·L−1 |
| KH2PO4 | 0.136 | g·L−1 |
| MgCl2·6H2O | 0.1017 | g·L−1 |
| CaCl2 | 0.055 | g·L−1 |
| 维生素溶液 | 1 | mL·L−1 |
| 微量元素溶液 | 1 | mL·L−1 |
| 超纯水 | 1 | L·L−1 |
| pH | 7.0 | ‒ |
表1 基础盐培养基组分
Table 1 The component of medium used in this study
| 组分 | 量值 | 单位 |
|---|---|---|
| 1,4-哌嗪二乙磺酸 (PIPES) | 3.024 | g·L−1 |
| NH4Cl | 0.535 | g·L−1 |
| NaHCO3 | 0.420 | g·L−1 |
| KH2PO4 | 0.136 | g·L−1 |
| MgCl2·6H2O | 0.1017 | g·L−1 |
| CaCl2 | 0.055 | g·L−1 |
| 维生素溶液 | 1 | mL·L−1 |
| 微量元素溶液 | 1 | mL·L−1 |
| 超纯水 | 1 | L·L−1 |
| pH | 7.0 | ‒ |
| 目的基因 | 引物名称 | 引物序列 (5'-3') | 长度/bp | 扩增程序 | 参考文献 |
|---|---|---|---|---|---|
| 细菌16S rRNA | 515F | GTGYCAGCMGCCGCGGTAA | 280 | 95 ℃ 30 s; 94 ℃ 20 s, 55 ℃ 20 s, 72 ℃ 30 s, 40个循环 | Einen et al., |
| 806R | GGACTACNVGGGTWTCTAAT | ||||
| 古菌16S rRNA | Arch519F | CAGCCGCCGCGGTAA | 400 | 95 ℃ 30 s; 95 ℃ 30 s, 57 ℃ 30 s, 72 ℃ 50 s, 40个循环 | Coolen et al., |
| Arch915R | GTGCTCCCCCGCCAATTCCT | ||||
| arsM | arsMF1 | TCYCTCGGCTGCGGCAAYCCVAC | 350 | 95 ℃ 30 s; 95 ℃ 30 s, 57 ℃ 30 s, 72 ℃ 1 min, 40个循环 | Jia et al., |
| arsMR2 | CGWCCGCCWGGCTTWAGYACCCG | ||||
| mcrA | mlas-mod-F | GGYGGTGTMGGDTTCACMCARTA | 469 | 95 ℃ 30 s; 95 ℃ 15 s, 58 ℃ 30 s, 72 ℃ 30 s, 40个循环 | Angel et al., |
| mcrA-rev-R | CGTTCATBGCGTAGTTVGGRTAGT |
表2 实时荧光定量PCR分析所用引物及扩增条件
Table 2 Lists of primer pairs and thermal cycling parameters for real-time quantitative PCR
| 目的基因 | 引物名称 | 引物序列 (5'-3') | 长度/bp | 扩增程序 | 参考文献 |
|---|---|---|---|---|---|
| 细菌16S rRNA | 515F | GTGYCAGCMGCCGCGGTAA | 280 | 95 ℃ 30 s; 94 ℃ 20 s, 55 ℃ 20 s, 72 ℃ 30 s, 40个循环 | Einen et al., |
| 806R | GGACTACNVGGGTWTCTAAT | ||||
| 古菌16S rRNA | Arch519F | CAGCCGCCGCGGTAA | 400 | 95 ℃ 30 s; 95 ℃ 30 s, 57 ℃ 30 s, 72 ℃ 50 s, 40个循环 | Coolen et al., |
| Arch915R | GTGCTCCCCCGCCAATTCCT | ||||
| arsM | arsMF1 | TCYCTCGGCTGCGGCAAYCCVAC | 350 | 95 ℃ 30 s; 95 ℃ 30 s, 57 ℃ 30 s, 72 ℃ 1 min, 40个循环 | Jia et al., |
| arsMR2 | CGWCCGCCWGGCTTWAGYACCCG | ||||
| mcrA | mlas-mod-F | GGYGGTGTMGGDTTCACMCARTA | 469 | 95 ℃ 30 s; 95 ℃ 15 s, 58 ℃ 30 s, 72 ℃ 30 s, 40个循环 | Angel et al., |
| mcrA-rev-R | CGTTCATBGCGTAGTTVGGRTAGT |
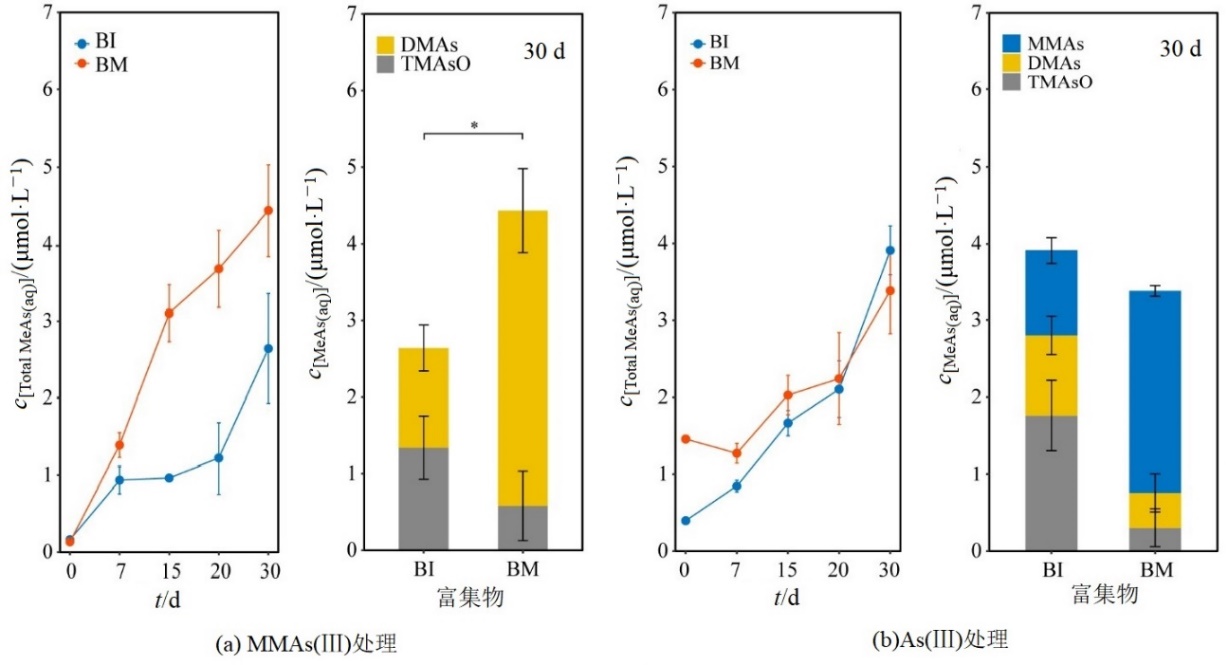
图1 BI和BM富集物中甲基砷物质的量浓度随时间变化和物种组成(以单位生物量计) *:P<0.05。下同
Figure 1 Evolution of methylated arsenic concentration and the composition of methylated arsenic species in BI and BM (normalized by OD600)
| [1] | ANGEL R, CLAUS P, CONRAD R, 2012. Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions[J]. The ISME Journal, 6(4): 847-862. |
| [2] |
BOLYEN E, RIDEOUT J R, DILLON M R, et al., 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2[J]. Nature Biotechnology, 37: 852-857.
DOI PMID |
| [3] |
BRAMMER H, RAVENSCROFT P, 2009. Arsenic in groundwater: A threat to sustainable agriculture in South and South-east Asia[J]. Environment International, 35(3): 647-654.
DOI PMID |
| [4] |
BRUCE R A, ACHENBACH L A, COATES J D, 1999. Reduction of (per)chlorate by a novel organism isolated from paper mill waste[J]. Environmental Microbiology, 1(4): 319-329.
DOI PMID |
| [5] |
CALLAHAN B J, MCMURDIE P J, ROSEN M J, et al., 2016. DADA2: High-resolution sample inference from Illumina amplicon data[J]. Nature Methods, 13: 581-583.
DOI PMID |
| [6] | CHEN C, LI L Y, WANG Y F, et al., 2023. Methylotrophic methanogens and bacteria synergistically demethylate dimethylarsenate in paddy soil and alleviate rice straighthead disease[J]. The ISME Journal, 17(11): 1851-1861. |
| [7] |
CHEN J, YOSHINAGA M, ROSEN B P, 2019. The antibiotic action of methylarsenite is an emergent property of microbial communities[J]. Molecular Microbiology, 111(2): 487-494.
DOI PMID |
| [8] | CHEN S C, SUN G X, YAN Y, et al., 2020. The Great Oxidation Event expanded the genetic repertoire of arsenic metabolism and cycling[J]. Proceedings of the National Academy of Sciences of the United States of America, 117(19): 10414-10421. |
| [9] | COOLEN M J L, HOPMANS E C, RIJPSTRA W I C, et al., 2004. Evolution of the methane cycle in Ace Lake (Antarctica) during the Holocene: response of methanogens and methanotrophs to environmental change[J]. Organic Geochemistry, 35(10): 1151-1167. |
| [10] | EINEN J, THORSETH I H, ØVREÅS L, 2008. Enumeration of Archaea and Bacteria in seafloor basalt using real-time quantitative PCR and fluorescence microscopy[J]. FEMS Microbiology Letters, 282(2): 182-187. |
| [11] |
GLISSMANN K, CONRAD R, 2000. Fermentation pattern of methanogenic degradation of rice straw in anoxic paddy soil[J]. FEMS Microbiology Ecology, 31(2): 117-126.
PMID |
| [12] | GROENIGEN K J V, KESSEL C V, HUNGATE B A, 2013. Increased greenhouse-gas intensity of rice production under future atmospheric conditions[J]. Nature Climate Change, 3: 288-291. |
| [13] | HATAMOTO M, IMACHI H, FUKAYO S, et al., 2007. Syntrophomonas palmitatica sp. nov., an anaerobic, syntrophic, long-chain fatty-acid-oxidizing bacterium isolated from methanogenic sludge[J]. International Journal of Systematic and Evolutionary Microbiology, 57(9): 2137-2142. |
| [14] |
HUANG K, LIU W, ZHAO F J, 2022. Methylarsenite is a broad-spectrum antibiotic disrupting cell wall biosynthesis and cell membrane potential[J]. Environmental Microbiology, 25(2): 562-574.
DOI PMID |
| [15] | HUANG K, XU Y, PACKIANATHAN C, et al., 2017. Arsenic methylation by a novel ArsM As(III) S-adenosylmethionine methyltransferase that requires only two conserved cysteine residues[J]. Molecular Microbiology, 107(2): 265-276. |
| [16] | JIA Y, HUANG H, ZHONG M, et al., 2013. Microbial arsenic methylation in soil and rice rhizosphere[J]. Environmental Science & Technology, 47(7): 3141-3148. |
| [17] |
KUMAR S, STECHER G, LI M, et al., 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Molecular Biology and Evolution, 35(6): 1547-1549.
DOI PMID |
| [18] |
LIU P F, QIU Q F, LU Y H, 2011. Syntrophomonadaceae-affiliated species as active butyrate-utilizing syntrophs in paddy field soil[J]. Applied and Environmental Microbiology, 77(11): 3884-3887.
DOI PMID |
| [19] | MASSCHELEYN P H, DELAUNE R D, PATRICK W H, 1991. Effect of redox potential and pH on arsenic speciation and solubility in a contaminated soil[J]. Environmental Science & Technology, 25(8): 1414-1419. |
| [20] | MCINERNEY M J, BRYANT M P, HESPELL R B, et al., 1981. Syntrophomonas wolfei gen. nov. sp. nov., an anaerobic, syntrophic, fatty acid-oxidizing bacterium[J]. Applied and Environmental Microbiology, 41(4): 1029-1039. |
| [21] |
MICHALKE K, WICKENHEISER E B, MEHRING M, et al., 2000. Production of volatile derivatives of metal(loid)s by microflora involved in anaerobic digestion of sewage sludge[J]. Applied and Environmental Microbiology, 66(7): 2791-2796.
DOI PMID |
| [22] | NILSSON R H, RYBERG M, KRISTIANSSON E, et al., 2006. Taxonomic reliability of DNA sequences in public sequence databases: A fungal perspective[J]. PLoS ONE, 1(1): e59. |
| [23] | QIAO J T, LI X M, HU M, et al., 2018. Transcriptional activity of arsenic-reducing bacteria and genes regulated by lactate and biochar during arsenic transformation in flooded paddy soil[J]. Environmental Science & Technology, 52(1): 61-70. |
| [24] | QIN J, ROSEN B P, ZHANG Y, et al., 2006. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase[J]. Proceedings of the National Academy of Sciences of the United States of America, 103(7): 2075-2080. |
| [25] | QUAST C, PRUESSE E, YILMAZ P, et al., 2013. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools[J]. Nucleic Acids Research, 41(D1): D590-D596. |
| [26] | REID M C, MAILLARD J, BAGNOUD A, et al., 2017. Arsenic methylation dynamics in a rice paddy soil anaerobic enrichment culture[J]. Environmental Science & Technology, 51(18): 10546-10554. |
| [27] | ROGNES T, FLOURI T, NICHOLS B, et al., 2016. VSEARCH: A versatile open source tool for metagenomics[J]. PeerJ, 4: e2584. |
| [28] |
RUI J P, PENG J J, LU Y H, 2009. Succession of bacterial populations during plant residue decomposition in rice field soil[J]. Applied and Environmental Microbiology, 75(14): 4879-4886.
DOI PMID |
| [29] |
SCHINK B, 1997. Energetics of syntrophic cooperation in methanogenic degradation[J]. Microbiology and Molecular Biology Reviews, 61(2): 262-280.
DOI PMID |
| [30] | SEGATA N, IZARD J, WALDRON L, et al., 2011. Metagenomic biomarker discovery and explanation[J]. Genome Biology, 12(6): R60. |
| [31] | SIERRA-ALVAREZ R, CORTINAS I, YENAL U, et al., 2004. Methanogenic inhibition by arsenic compounds[J]. Applied and Environmental Microbiology, 70(9): 5688-5691. |
| [32] |
STAMS A J M, PLUGGE C M, 2009. Electron transfer in syntrophic communities of anaerobic bacteria and archaea[J]. Nature Reviews Microbiology, 7: 568-577.
DOI PMID |
| [33] | VIACAVA K, MEIBOM K L, ORTEGA D, et al., 2020. Variability in arsenic methylation efficiency across aerobic and anaerobic microorganisms[J]. Environmental Science & Technology, 54(22): 14343-14351. |
| [34] | VIACAVA K, QIAO J T, JANOWCZYK A, et al., 2022. Meta-omics-aided isolation of an elusive anaerobic arsenic-methylating soil bacterium[J]. The ISME Journal, 16(7): 1740-1749. |
| [35] | VRIENS B, LENZ M, CHARLET L, et al., 2014. Natural wetland emissions of methylated trace elements[J]. Nature Communications, 5: 3035/1-3035/7. |
| [36] | WANG P P, SUN G X, ZHU Y G, 2014. Identification and characterization of arsenite methyltransferase from an archaeon, Methanosarcina acetivorans C2A[J]. Environmental Science & Technology, 48(21): 12706-12713. |
| [37] | WEBSTER T M, REDDY R R, TAN J Y, et al., 2016. Anaerobic disposal of arsenic-bearing wastes results in low microbially mediated arsenic volatilization[J]. Environmental Science & Technology, 50(20): 10951-10959. |
| [38] | WOLIN E A, WOLFE R S, WOLIN M J, 1964. Viologen dye inhibition of methane formation by Methanobacillus omelianskii [J]. Journal of Bacteriology, 87(5): 993-998. |
| [39] | YU G C, SMITH D K, ZHU H C, et al., 2017. GGtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data[J]. Methods in Ecology and Evolution, 8: 28-36. |
| [40] | ZHAO F J, HARRIS E, YAN J, et al., 2013. Arsenic methylation in soils and its relationship with microbial arsM abundance and diversity, and as speciation in rice[J]. Environmental Science & Technology, 47(13): 7147-7154. |
| [41] |
ZHU Y G, YOSHINAGA M, ZHAO F J, et al., 2014. Earth abides arsenic biotransformations[J]. Annual Review of Earth and Planetary Sciences, 42: 443-467.
DOI |
| [42] |
ZOU B Z, TAKEDA K, TONOUCHI A, et al., 2003. Characteristics of an anaerobic, syntrophic, butyrate-degrading bacterium in paddy field soil[J]. Bioscience, Biotechnology, and Biochemistry, 67(10): 2059-2067.
PMID |
| [43] | 费媛媛, 焦硕, 陆雅海, 2021. 中国东部水稻土壤丁酸互营降解微生物的地理分布格局[J]. 北京大学学报(自然科学版), 57(1): 143-152. |
| FEI Y Y, JIAO S, LU Y H, 2021. Biogeographic patterns of microbial communities associated with syntrophic butyrate degradation in paddy soils in eastern China[J]. Acta Scientiarum Naturalium Universitatis Pekinensis, 57(1): 143-152. | |
| [44] |
胡启武, 吴琴, 刘影, 等, 2009. 湿地碳循环研究综述[J]. 生态环境学报, 18(6): 2381-2386.
DOI |
| HU Q W, WU Q, LIU Y, et al., 2009. A review of carbon cycle in wetlands[J]. Ecology and Environmental Sciences, 18(6): 2381-2386. | |
| [45] | 李令仪, 张楠, 张洋, 等, 2023. 稻田土壤可溶性有机碳荧光组分对砷甲基化的影响[J]. 农业环境科学学报, 42(10): 2211-2219. |
| LI L Y, ZHANG N, ZHANG Y, et al., 2023. Effects of soluble organic carbon fluorescence components on arsenic methylation in paddy soil[J]. Journal of AgroEnvironment Science, 42(10): 2211-2219. | |
| [46] | 李晓敏, 牟山, 陈娅婷, 等, 2019. 稻田土壤微生物驱动的微好氧亚铁氧化耦合碳同化过程[J]. 中国科学:地球科学, 49(12): 1948-1959. |
| LI X M, MOU S, CHEN Y T, et al., 2019. Microaerobic Fe(II) oxidation coupled to carbon assimilation processes driven by microbes from paddy soil[J]. Science China Earth Sciences, 49(12): 1948-1959. | |
| [47] | 刘鹏飞, 陆雅海, 2013. 水稻土中脂肪酸互营氧化的研究进展[J]. 微生物学通报, 40(1): 109-122. |
| LIU P F, LU Y H, 2013. A review of syntrophic fatty acids oxidation in anoxic paddy soil[J]. Microbiology China, 40(1): 109-122. | |
| [48] |
邱丽娟, 吴攀, 张翅鹏, 等, 2018. 高砷煤矿污染稻田水稻对砷的吸收与赋存特征研究[J]. 生态环境学报, 27(7): 1292-1297.
DOI |
| QIU L J, WU P, ZHANG C P, et al., 2018. Study on the uptake and occurrence features of arsenic in rice in the field contaminated by high arsenic coal mine[J]. Ecology and Environmental Sciences, 27(7): 1292-1297. | |
| [49] | 田腾, 颜蒙蒙, 曾希柏, 等, 2020. 不同来源可溶性有机质对稻田土壤中砷甲基化的影响[J]. 农业环境科学学报, 39(3): 511-520. |
| TIAN T, YAN M M, ZENG X B, et al., 2020. Effect of dissolved organic matter from different sources on arsenic methylation in paddy soils[J]. Journal of Agro-Environment Science, 39(3): 511-520. | |
| [50] | 王培培, 陈松灿, 朱永官, 等, 2018. 微生物砷甲基化及挥发研究进展[J]. 农业环境科学学报, 37(7): 1377-1385. |
| WANG P P, CHEN S C, ZHU Y G, et al., 2018. Advances in the research of arsenic methylation and volatilization by microorganisms[J]. Journal of Agro-Environment Science, 37(7): 1377-1385. | |
| [51] | 张杰, 陆雅海, 2015. 互营氧化产甲烷微生物种间电子传递研究进展[J]. 微生物学通报, 42(5): 920-927. |
| ZHANG J, LU Y H, 2015. A review of interspecies electron transfer in syntrophic-methanogenic associations[J]. Microbiology China, 42(5): 920-927. |
| [1] | 杨宇, 邓仁健, 隆佩, 黄中杰, 任伯帜, 王政华. 砷氧化菌Pseudomonas sp. AO-1的分离鉴定及其对As(Ⅲ)的氧化性能研究[J]. 生态环境学报, 2023, 32(3): 619-626. |
| [2] | 尹浩均, 龙明亮, 刘维, 倪春林, 李芳柏, 吴云当. 溶氧浓度调控嗜水气单胞菌的砷还原:效应与机制[J]. 生态环境学报, 2023, 32(2): 381-387. |
| [3] | 董乐恒, 王旭刚, 陈曼佳, 王子豪, 孙丽蓉, 石兆勇, 吴琪琪. 光照和避光条件下石灰性水稻土Fe氧化还原与Cu活性关系研究[J]. 生态环境学报, 2022, 31(7): 1448-1455. |
| [4] | 张亚平, 陈慧敏, 吴志宇, 汤佳, 谢章彰, 刘芳华. 低量水铁矿促进稻田梭菌Clostridium sp. BY-1产氢效率[J]. 生态环境学报, 2022, 31(12): 2341-2349. |
| [5] | 黄成, 吴月颖, 吉恒宽, 陈丽铭, 李倍莹, 符传良, 李建宏, 吴蔚东, 吴治澎. 海南典型水稻土厌氧铁还原特征对DOM分子特性的响应[J]. 生态环境学报, 2021, 30(5): 957-967. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
